Overview of the First Interactive Prototype
The Prototype
Our interactive prototype relies heavily on the production version of IMG, since implementing even a small portion of the data analysis tools made available by IMG is beyond the scope of this course. In a real production version of IMG running Geneboree collaborative annotation, all the pages would be served by one server, and the whole would behave as one seamless system.
The line of separation between servers reflects the line of separation between functionality in IMG that we have not attempted to redesign (the searching and comparative analysis tools) and the new functionality we are developing (annotation, communication between users about gene function assignments, and authentication).
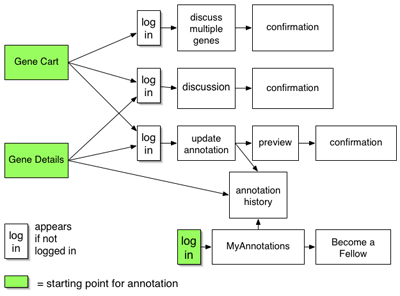
Interaction flow through the IMG system with geneboree collaborative annotation
Our approach is to allow users to tap into the wealth of genomics tools made available by the existing system in order to determine how best to annotate the genes in which they are interested. It is therefore crucial for our users to have ready access to pages lke the gene details page, searches, and the phylogenetic profiler while filling in annotation data. We allow this by opening the form for updating an annotation in a popup window.
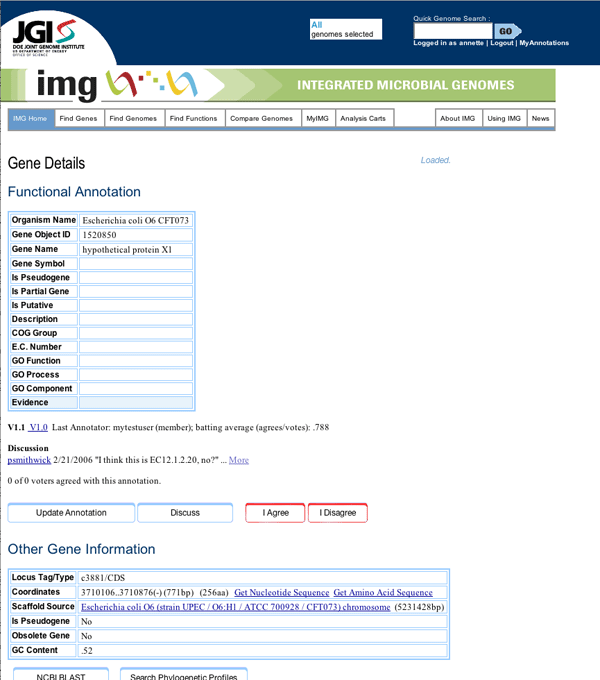
Top portion of the Gene Details page showing the annotation area
Inside the popup window, we provide three major sections, one for entering de novo information, one for transferring information from an already-annotated gene likely to have the same function, and one for recording the evidence that led the annotator to their conclusions.
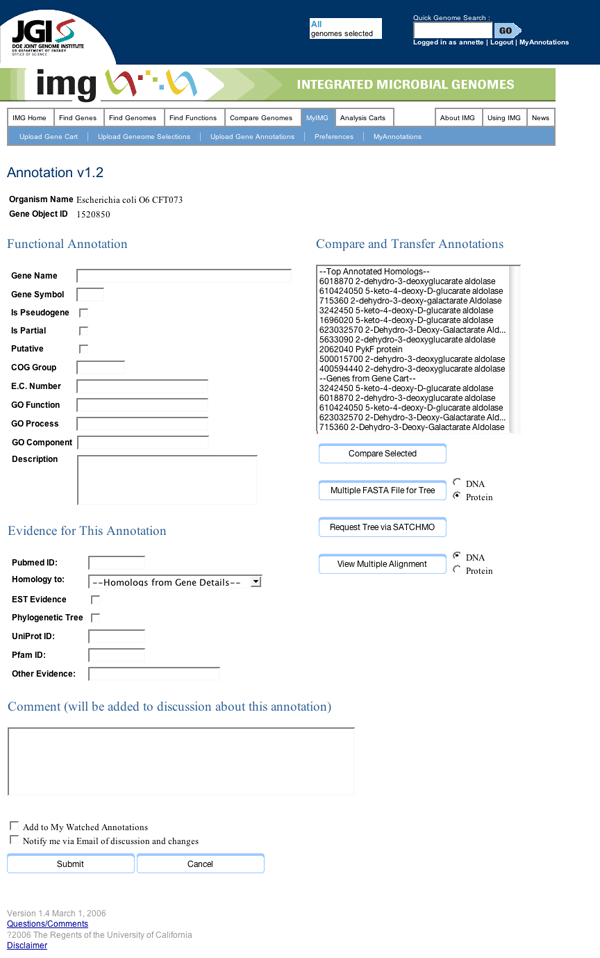
Update Annotation page
One can reach the annotation popup via a link on the gene details page for a particular gene, in this case a 2-dehydro-3-deoxyglucarate aldolase gene we have renamed "hypothetical protein X1", or from the gene cart. Gene carts are an existing feature of IMG where users place genes in an online shopping cart as they browse the site. From the cart, users can run comparisons on sets of genes. This "select some genes, put them in your cart, run comparisons" workflow is the central workflow of the IMG system, and IMG users are familiar with it.
Our prototype implements two methods of communicating between collaborators, discussions and voting. In discussions, annotators may query each other about why certain assignments were made, suggest hypotheses they would not feel comfortable entering as regular annotations, and generally collaborate. Our system allows users to enter discussions about the current versions of the functional annotation for one or many genes.
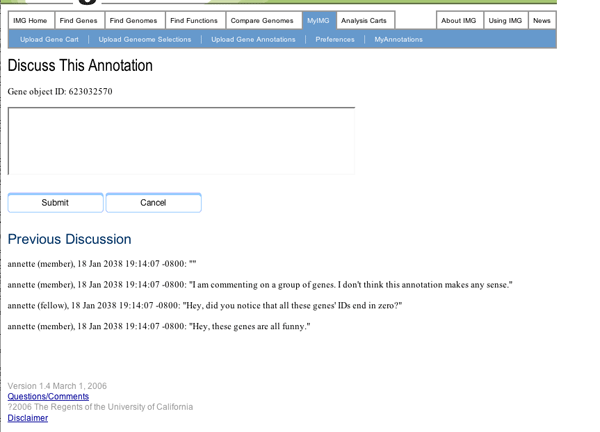
Page for entering a comment as part of the discussion about a gene's current annotation
The voting system allows users who don't care to spend time annotating or discussing a given gene to simply indicate their agreement or disagreement with the current version of an annotation. We chose to keep the voting options simple, because we didn't want users to get slowed down by trying to evaluate the quality of an annotation. In discussions with users, we observed that the typical response to an annotation is quite binary, either the scientist feels the assignment is accurate or they feel that it isn't. Where they seek fine-grained data is in assessing the reliability of the assignment. This usually means knowing the source (was the annotation automated? manual? done by an investigator with a good reputation?) and what evidence the source used (such as a publication, homology to another gene whose function is known, or analyzing a phylogenetic tree). The voting results are used to show a ratio that essentially rates the consensus of opinion about a given version of an annotation. The ratios are in turn used to generate a "batting average" for a given annotator. The batting average is the overall ratio of agrees to votes for all of an annotator's annotations. Users felt that a star-type rating of annotators would seem overly judgmental, but they responded very positively to the batting average concept.
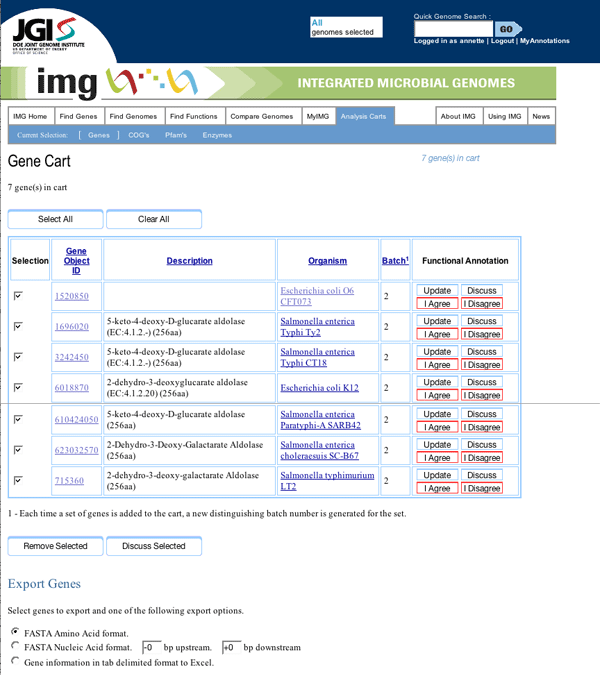
Gene Cart page with voting buttons
To address security and reliability issues, our system allows two types of users. Any individual can sign up as a member, which allows them to update annotations but does not allow them to vote or perform batch operations (i.e., entering discussion comments about multiple genes or transferring all annotation data from one gene to another with a single click). Users who have entered more than 10 annotations and kept a batting average above .500 are automatically made "fellows". Besides being able to vote and run batch operations, Existing fellows also have the right to make other users fellows, giving trusted individuals full access right away. When one is browsing as a fellow, usernames of annotators also appear as mailto links. In our prototype, all users have fellow-level access.
What We Left Out
There are a few small items missing from the update area, simply because we did not have time to implement them. One is the ability to enter a second pubmed ID as evidence for an annotation. Another is the appearance of corresponding ontology terms beside number entry fields, so that annotators can verify that they have entered the right number. There is also no form validation as yet. The Compare and Transfer page is missing a javascript behavior that will select all the radio buttons for a given homolog when the user elects to transfer all fields from that gene. It also does not actually transfer data to the update page as it should.
We still plan to design and implement a method for replying to discussions about multiple genes so that the reply is automatically assigned to all the genes in the group if that is what the user desires. We chose to leave this out because it is not required in our scenarios.
Finally, as another time saver, not all data is currently pulled from the database. Some pages have hard-coded data that will not update when new data is submitted to the system. For example, the batting averages are not currently calculated, and new annotations are not actually saved.
Tools
We used Dreamweaver with its template system to control the aspects of our system that are the same on multiple pages. To enable dynamic functionality, we used PHP and a MySQL database. Our Dreamweaver templates are nested, allowing changes to the entire site or to subsets of pages without affecting the site as a whole. This should allow for relatively rapid iteration of changes. Unfortunately, this occasionally complicates things, when Dreamweaver finds a tag that spans the border between template code and non-template code. This can arise even in clean code when a tag is opened outside PHP and closed inside PHP. Getting up to speed with PHP was rather time-consuming, as not all our project team had used it, and only one had used it recently. We chose to use PHP because, among the scripting languages that at least some of us knew, it offered the best ease of learning and remembering.