The JGI Genome Portal
The Geneboree annotation system for the Integrated Microbial Genomes (IMG) system has as its closest analog the existing functional annotation tool currently available on the Joint Genome Institute (JGI) “Genome Portal.” The JGI core business model is to provide high-quality sequencing and related services for the scientific community. Gene prediction and functional annotation based on prediction algorithms are two such services, but JGI's annotation group no longer does manual annotation. Instead, it trains a selected set of collaborators to manually annotate genomes that have not yet been publicly released. (With the Human Genome Project now complete, the institute’s focus has shifted to extending genome sequencing to a diverse array of organisms.) Once finished and annotated by collaborators, a genome is made available publicly on the Genome Portal and is loaded into IMG as well. The IMG system incorporates all of the microbial data from JGI’s Genome Portal but has a broader scope: it seeks to build a data management platform for a collection of sequences from various sources, including sequencing centers worldwide.
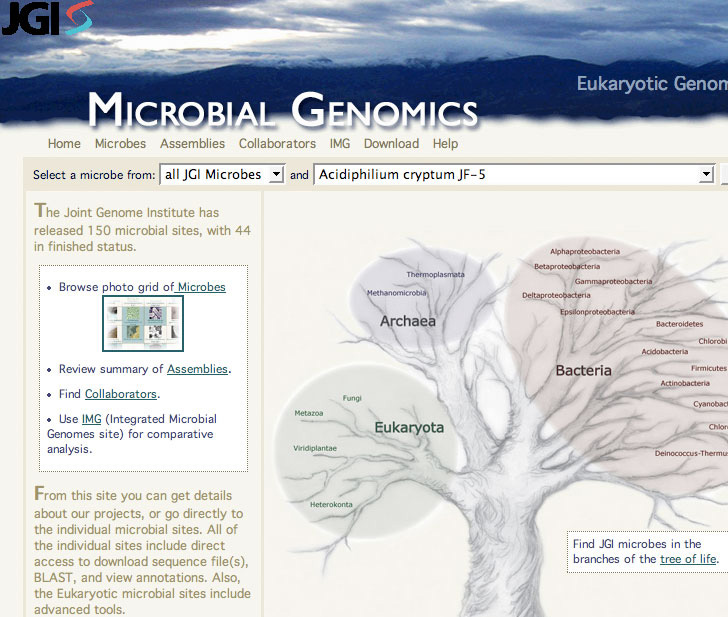
The Genome Portal does not have a single annotation area. Rather, resources are organized by species under two subsites, reached via links labeled either Eukaryotic Genomics or Microbial Genomics. Sites for each of the organisms of interest can be accessed via alphabetized drop-down menus found on each of the two sub-sites. The Genome Portal also includes a “tree of life” imagemap that provides links to sites for specific organisms. Based on a tree metaphor, it attempts to depict lineage on the branches of a tree drawing. It seems to be counter-intuitive for scientific purposes and difficult to navigate (Fig 1).
Figure 1:Tree of Life Image Map, used to navigate to specific organisms.
The Microbial Genomics collection includes 44 finished sequences and 90 drafts. Every organism has a home page that provides access to comparative analysis tools that one might need when making a manual functional or structural assignment. Tools for BLAST, GO, KEGG, COG, and searching are provided for finished eukaryotic genomes. A genome browser is available under the “Browse” tab. There is also access to a “Help” section and an option to download the complete sequence in FASTA format (Fig 2).
Figure 2: The Microbial Genetics Genome Browser.

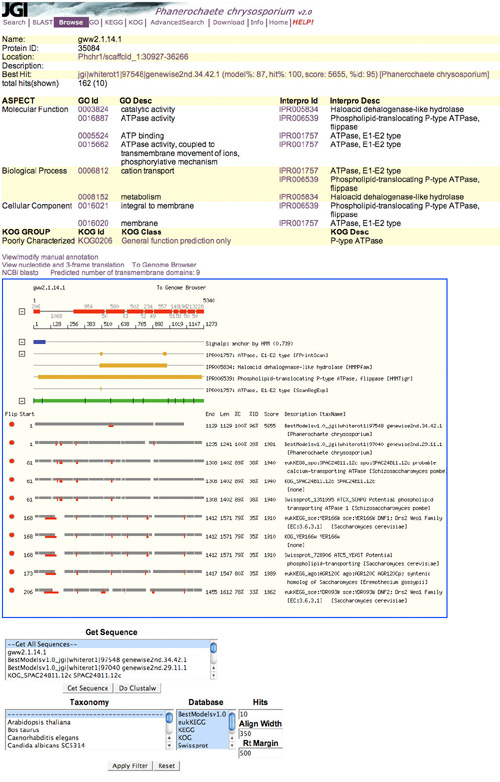
To annotate a genome, users identify a gene model, either through the browser or via one of the other tools. The genome browser displays models within tracks (colored, horizontal rows of sequence data) representing the scaffold, JGI-predicted genes ("filtered gene models"), and other relevant sequences (such as models predicted by specific algorithms), all aligned by base position. Each track is shown in a specific color and with an identifying label. For an organism under active annotation, the "catalog" track constitutes the JGI reference list. Structural annotation consists mostly of deciding whether each model in the catalog is correct, and if not, removing it from the catalog and promoting an alternative gene model. Functional annotation from the browser requires navigating to a gene model, clicking on it to view a summary page about the gene, and finding and clicking on a very small "view/modify manual annotation" link.
Track labels are a little cryptic and not explicitly attributed. The number of filtered models is usually large and runs off the page. When the user clicks on a specific gene in one of the tracks and then returns to the browser, it doesn’t put him back at the same track. Instead, it reloads the page from the top and the user has to scroll down to find the track he has been working on. Overall, creating or annotating one gene model is a tedious process that requires a lot of clicking.
One can also use the Advanced Search tool to find a gene model. In the results, one finds a series of letter-coded links beside each hit. Clicking on the T brings up the annotation page. The Advanced Search page searches only the gene catalog and filtered model tracks. The simple search page allows users to search all models. It can also be used to search the annotations of models with hits to protein databases. One can narrow the search to models within a specific track and search by an ID. The GO, KEGG, and COG tools can also be used for generating lists of gene models that have been automatically assigned to a pathway or function.
Specific genes' pages (Fig. 3) are accessed by clicking on the gene on in the genome browser or in the result list from another tool. These pages display all of the relevant information about a specific gene. If the gene's functional assignment has been edited manually, the gene page also shows the user name of the annotator and his description. Unfortunately, it does not provide a simple way to contact the user responsible for the annotation (a link to their email address, for example). The high-scoring alignments are shown graphically at the bottom.
Figure 3: Gene Details Page
The design of the annotation page (Fig. 4) is inconsistent with the rest of the site. Buttons for editing or adding annotations become accessible once the user logs in. Tiny “edit” or “add” links appear on each row (Fig. 5). Each value can be modified by clicking on the link, calling up a separate form in a pop-up window (Fig. 6). The approach seems tedious and could be improved by converting it to a simpler form.
Figure 4: JGI's Annotation Page
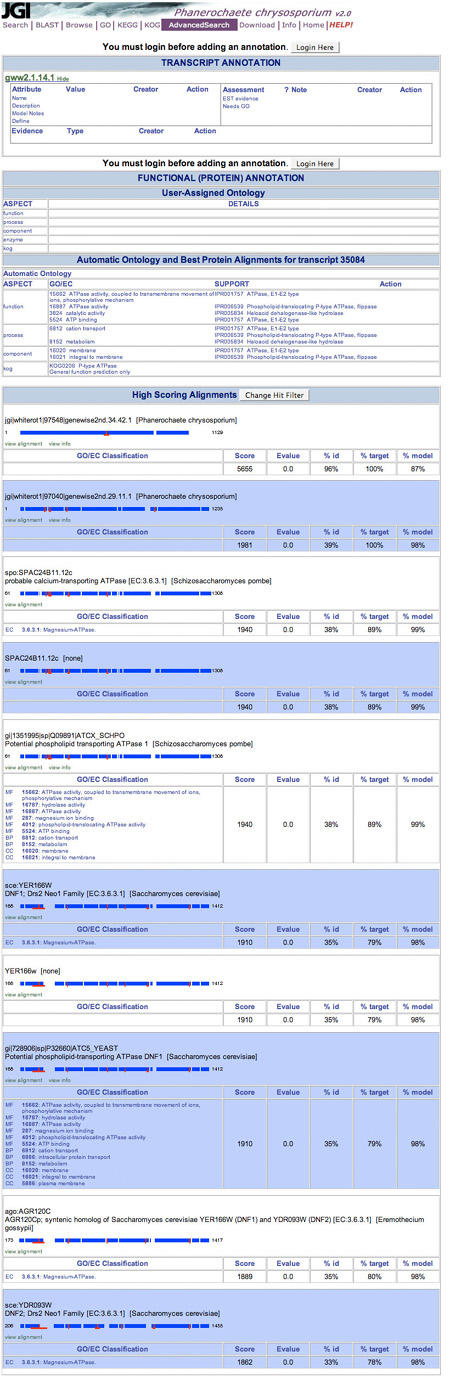
Figure 5: Detail of Annotation Modification Interface

Figure 6: Pop-Up Screen. This pop-up must be used for modification of each individual field.

Overall, JGI's Genome Portal site includes a vast array of essential resources for performing comparative manual annotation. The site is complex and its users usually go through tutorial sessions in order to learn how to annotate.