The Entrez Gene Database
RefSeq, the Reference Sequence collection, aims to provide a comprehensive, integrated, non-redundant set of sequences, including genomic DNA, transcript (RNA), and protein products, for major research organisms. The Entrez Gene database is a searchable database of genes from RefSeq genomes. Of interest to us is the GeneRIF feature of this database – short for Gene Reference Into Function. GeneRIF provides a simple mechanism to allow scientists to add to the functional annotation of genes described in Entrez Gene. Three types of information are required to complete a submission:
- A concise phrase describing a function or functions (less than 255 characters in length, preferably more than a restatement of the title of the paper).
- A published paper describing that function, implemented by supplying the PubMed ID of a citation in PubMed.
- A valid e-mail address (which will remain confidential).
Access to the submission form is provided from the Bibliography section of the Entrez Gene default report view, so you don't have to retype the Gene ID or other identifiers. GeneRIFs are intended to facilitate access to publications documenting experiments that add to our understanding of a gene and its function. Reports based solely on computational analyses are not in scope.
Figure 1: Entrez Gene Search Interface
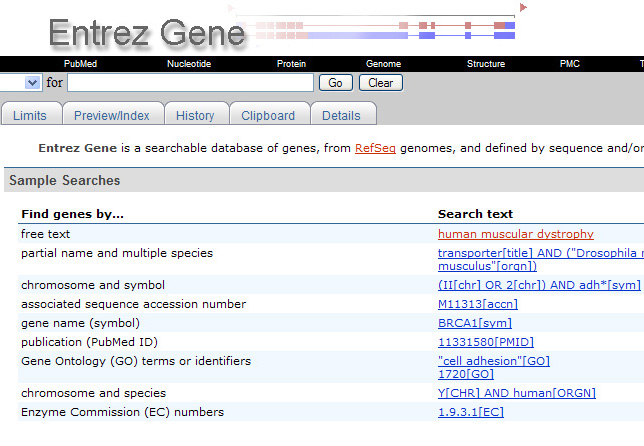
Figure 2: GeneRIF Submission Form
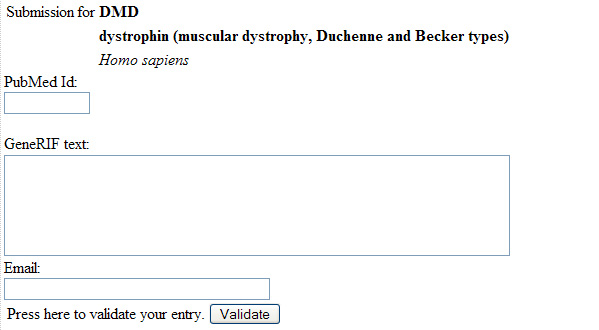
Figure 3: GeneRif Modification Form
Figure 4: Entrez Search Results, Including GeneRIF*

*note that Summary, Genomic Regions, and Genomic Context information have been truncated in this image, in order to show regions of interest.
Strengths
- Versatile gene search interface
- Simple GeneRIF upload mechanism
- Many genes have a good-sized, reliable, manually-verified set of annotations.
Weaknesses
- Difficult to correct or modify existing GeneRIFs. One must navigate to a completely distinct form and enter data without type-checking.
- Sometimes the sheer number of GeneRIFs can be confusing.
- No comparative sense of how important each GeneRIF is. Order appears to be based on chronology of submission.
Lessons Learned
We have much to learn from the Entrez database, in light of the fact that it is one of the most reputed and content-rich sources of gene annotation available today. For instance, it draws an important distinction between annotations derived from publications and those derived from computational analysis: that the latter is far from reliable (and generally not even accepted). One wonders, though, if there exists a way to make use of the computational analysis data without discrediting the body of annotation information altogether. Another nice feature is the ease of submission and transparency of authorship, although one might envision a better way of managing revisions/modifications about a certain annotation than through the bottleneck of a single, original author. Lastly, upon returning the search results to a given query, the GeneRIFs are displayed in a clear, easily understood manner - although it would be instructive to see whether there is a better way to organize or group such information.