Interview with Jane Doe 1 and Jane Doe 2 at JGI
Have you used JGI's Integrated Microbial Genomes system?
All the time.
• What role does manual annotation play in your work?
None anymore. It was part of their regular work when JGI was still sequencing their section of the human genome, but now that they are done, they leave all annotation to off-site collaborators.
• Breakdown of work area: What's what? Explanation of physical workspace structure. Looking for piles of paper, desk structure, communal vs. personal, etc.
The main resources they used to use when annotating were web sites.
HumanPsych, GenBank (RefSeek), KEG, GO, COG
• Please show us the process of doing a manual annotation. (Identify tasks. Can you break down your main task into a series of small sub-tasks?)
There are a couple of ways to do manual annotation. One is that you are assigned a particular set of the chromosome (locus annotation), and you work on subsequent chunks of chromosomes. Another way is to focus on pathways or related functions, which will annotate disparate sets of chromosomes.
We were walked through the JGI website current annotation tool, which has a lot of very small print, 'add' icons, and pop-up windows. There is a space to indicate action items, including "GO annotation required." Various technical terms are explained.
Annotations or changes in annotations often cite an authoritative reference, such as a PubMed ID.
• What information do you need in order to do an annotation? What resources do you currently use while annotating? (include anything from google to post-it notes...)
You draw a lot of information from the whole JGI network, shown below.
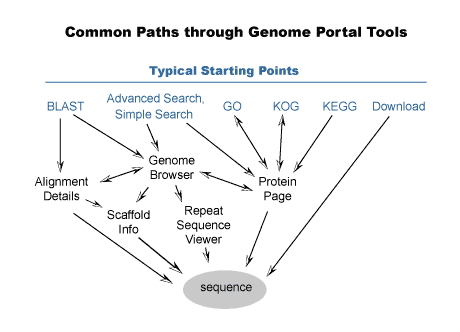
• What role does collaboration with other annotators play in your work? If you do talk to other annotators, what do you communicate with the other annotators about? Do you talk to them all at once?
There are debates, for example, on the proper name to associate with a particular gene. When the conflict cannot be resolved, sometimes the gene is given two names. At the time when they were doing manual annotation, there was a distributed team of 11 or so annotators, communicating over web-based forms, email, and sometimes instant messenger.
If no name is assigned, a unique identified is generated automatically.
• Would you want to share/be able to see other people's work in progress (tentative annotations)?
The interviewees went on a bit of a tangent on the current existing interface and how it links annotations to the annotators. She said the communities are very tight, such that you would even be able to recognize the alias of the annotator immediately. The implication here is that the reason the communities are so tight is because there is so much informal dialogue happening.
• How do you know when you have done enough annotation on a genome to call it a new version?
The basic requirement is:
A gene name.
A definition.
Other fields:
Description
Disposition
Model Notes
Fields that are typically annotated:
Name
Definition
Description
Disposition
Description
(Not Model)
She also explained the concept of CDNA and EST.